Deep Learning: Convolutional Neural Networks and Recurrent Neural Networks
Jan 13, 2023
Plans for January 14 and 15, 2023
- Saturday January 14: Convolutional Neural Networks (CNN)
- Sunday January 15: Recurrent Neural Networks (RNN)
- Video of lecture to be added
- Discussion of last project
Reading Recommendations
- Goodfellow, Bengio, Courville, chapter 9
- We will follow to a large extent the lectures from CS231 at Stanford
- Michael Nielsen's book is a must read, in particular chapter 6 which deals with CNNs.
- Goodfellow et al, chapter 10 on Recurrent NNs, chapters 11 and 12 on various practicalities around deep learning are also recommended.
- Lectures from CS231 at Stanford
- Aurelien Geron, chapter 14 on RNNs.
Convolutional Neural Networks (recognizing images)
Convolutional neural networks (CNNs) were developed during the last decade of the previous century, with a focus on character recognition tasks. Nowadays, CNNs are a central element in the spectacular success of deep learning methods. The success in for example image classifications have made them a central tool for most machine learning practitioners.
CNNs are very similar to ordinary Neural Networks. They are made up of neurons that have learnable weights and biases. Each neuron receives some inputs, performs a dot product and optionally follows it with a non-linearity. The whole network still expresses a single differentiable score function: from the raw image pixels on one end to class scores at the other. And they still have a loss function (for example Softmax) on the last (fully-connected) layer and all the tips/tricks we developed for learning regular Neural Networks still apply (back propagation, gradient descent etc etc).
What is the Difference
CNN architectures make the explicit assumption that the inputs are images, which allows us to encode certain properties into the architecture. These then make the forward function more efficient to implement and vastly reduce the amount of parameters in the network.
Here we provide only a superficial overview, for the more interested, we recommend highly the course IN5400 – Machine Learning for Image Analysis and the slides of CS231.
Another good read is the article here https://arxiv.org/pdf/1603.07285.pdf.
Neural Networks vs CNNs
Neural networks are defined as affine transformations, that is a vector is received as input and is multiplied with a matrix of so-called weights (our unknown paramters) to produce an output (to which a bias vector is usually added before passing the result through a nonlinear activation function). This is applicable to any type of input, be it an image, a sound clip or an unordered collection of features: whatever their dimensionality, their representation can always be flattened into a vector before the transformation.
Why CNNS for images, sound files, medical images from CT scans etc?
However, when we consider images, sound clips and many other similar kinds of data, these data have an intrinsic structure. More formally, they share these important properties:
- They are stored as multi-dimensional arrays (think of the pixels of a figure) .
- They feature one or more axes for which ordering matters (e.g., width and height axes for an image, time axis for a sound clip).
- One axis, called the channel axis, is used to access different views of the data (e.g., the red, green and blue channels of a color image, or the left and right channels of a stereo audio track).
These properties are not exploited when an affine transformation is applied; in fact, all the axes are treated in the same way and the topological information is not taken into account. Still, taking advantage of the implicit structure of the data may prove very handy in solving some tasks, like computer vision and speech recognition, and in these cases it would be best to preserve it. This is where discrete convolutions come into play.
A discrete convolution is a linear transformation that preserves this notion of ordering. It is sparse (only a few input units contribute to a given output unit) and reuses parameters (the same weights are applied to multiple locations in the input).
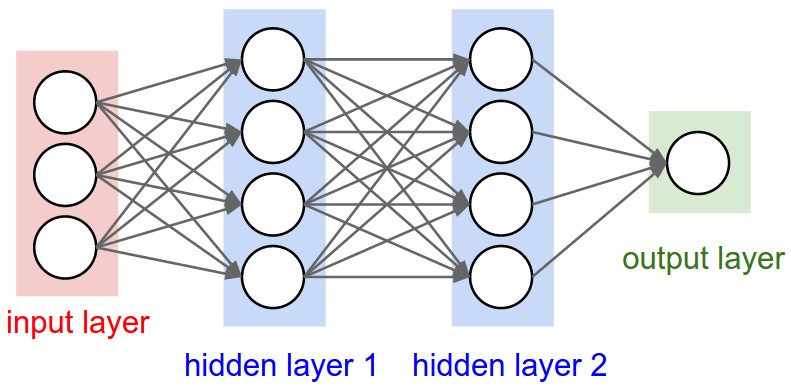
Regular NNs don’t scale well to full images
As an example, consider an image of size \( 32\times 32\times 3 \) (32 wide, 32 high, 3 color channels), so a single fully-connected neuron in a first hidden layer of a regular Neural Network would have \( 32\times 32\times 3 = 3072 \) weights. This amount still seems manageable, but clearly this fully-connected structure does not scale to larger images. For example, an image of more respectable size, say \( 200\times 200\times 3 \), would lead to neurons that have \( 200\times 200\times 3 = 120,000 \) weights.
We could have several such neurons, and the parameters would add up quickly! Clearly, this full connectivity is wasteful and the huge number of parameters would quickly lead to possible overfitting.
Figure 1: A regular 3-layer Neural Network.
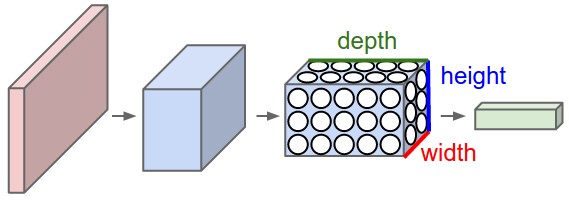
3D volumes of neurons
Convolutional Neural Networks take advantage of the fact that the input consists of images and they constrain the architecture in a more sensible way.
In particular, unlike a regular Neural Network, the layers of a CNN have neurons arranged in 3 dimensions: width, height, depth. (Note that the word depth here refers to the third dimension of an activation volume, not to the depth of a full Neural Network, which can refer to the total number of layers in a network.)
To understand it better, the above example of an image with an input volume of activations has dimensions \( 32\times 32\times 3 \) (width, height, depth respectively).
The neurons in a layer will only be connected to a small region of the layer before it, instead of all of the neurons in a fully-connected manner. Moreover, the final output layer could for this specific image have dimensions \( 1\times 1 \times 10 \), because by the end of the CNN architecture we will reduce the full image into a single vector of class scores, arranged along the depth dimension.
Figure 2: A CNN arranges its neurons in three dimensions (width, height, depth), as visualized in one of the layers. Every layer of a CNN transforms the 3D input volume to a 3D output volume of neuron activations. In this example, the red input layer holds the image, so its width and height would be the dimensions of the image, and the depth would be 3 (Red, Green, Blue channels).
Layers used to build CNNs
A simple CNN is a sequence of layers, and every layer of a CNN transforms one volume of activations to another through a differentiable function. We use three main types of layers to build CNN architectures: Convolutional Layer, Pooling Layer, and Fully-Connected Layer (exactly as seen in regular Neural Networks). We will stack these layers to form a full CNN architecture.
A simple CNN for image classification could have the architecture:
- INPUT (\( 32\times 32 \times 3 \)) will hold the raw pixel values of the image, in this case an image of width 32, height 32, and with three color channels R,G,B.
- CONV (convolutional )layer will compute the output of neurons that are connected to local regions in the input, each computing a dot product between their weights and a small region they are connected to in the input volume. This may result in volume such as \( [32\times 32\times 12] \) if we decided to use 12 filters.
- RELU layer will apply an elementwise activation function, such as the \( max(0,x) \) thresholding at zero. This leaves the size of the volume unchanged (\( [32\times 32\times 12] \)).
- POOL (pooling) layer will perform a downsampling operation along the spatial dimensions (width, height), resulting in volume such as \( [16\times 16\times 12] \).
- FC (i.e. fully-connected) layer will compute the class scores, resulting in volume of size \( [1\times 1\times 10] \), where each of the 10 numbers correspond to a class score, such as among the 10 categories of the MNIST images we considered above . As with ordinary Neural Networks and as the name implies, each neuron in this layer will be connected to all the numbers in the previous volume.
Transforming images
CNNs transform the original image layer by layer from the original pixel values to the final class scores.
Observe that some layers contain parameters and other don’t. In particular, the CNN layers perform transformations that are a function of not only the activations in the input volume, but also of the parameters (the weights and biases of the neurons). On the other hand, the RELU/POOL layers will implement a fixed function. The parameters in the CONV/FC layers will be trained with gradient descent so that the class scores that the CNN computes are consistent with the labels in the training set for each image.
CNNs in brief
In summary:
- A CNN architecture is in the simplest case a list of Layers that transform the image volume into an output volume (e.g. holding the class scores)
- There are a few distinct types of Layers (e.g. CONV/FC/RELU/POOL are by far the most popular)
- Each Layer accepts an input 3D volume and transforms it to an output 3D volume through a differentiable function
- Each Layer may or may not have parameters (e.g. CONV/FC do, RELU/POOL don’t)
- Each Layer may or may not have additional hyperparameters (e.g. CONV/FC/POOL do, RELU doesn’t)
For more material on convolutional networks, we strongly recommend the course IN5400 – Machine Learning for Image Analysis and the slides of CS231 which is taught at Stanford University (consistently ranked as one of the top computer science programs in the world). Michael Nielsen's book is a must read, in particular chapter 6 which deals with CNNs.
The textbook by Goodfellow et al, see chapter 9 contains an in depth discussion as well.
Key Idea
A dense neural network is representd by an affine operation (like matrix-matrix multiplication) where all parameters are included.
The key idea in CNNs for say imaging is that in images neighbor pixels tend to be related! So we connect only neighboring neurons in the input instead of connecting all with the first hidden layer.
We say we perform a filtering (convolution is the mathematical operation).
Mathematics of CNNs
The mathematics of CNNs is based on the mathematical operation of convolution. In mathematics (in particular in functional analysis), convolution is represented by mathematical operation (integration, summation etc) on two function in order to produce a third function that expresses how the shape of one gets modified by the other. Convolution has a plethora of applications in a variety of disciplines, spanning from statistics to signal processing, computer vision, solutions of differential equations,linear algebra, engineering, and yes, machine learning.
Mathematically, convolution is defined as follows (one-dimensional example): Let us define a continuous function \( y(t) \) given by
$$ y(t) = \int x(a) w(t-a) da, $$where \( x(a) \) represents a so-called input and \( w(t-a) \) is normally called the weight function or kernel.
The above integral is written in a more compact form as
$$ y(t) = \left(x * w\right)(t). $$The discretized version reads
$$ y(t) = \sum_{a=-\infty}^{a=\infty}x(a)w(t-a). $$Computing the inverse of the above convolution operations is known as deconvolution.
How can we use this? And what does it mean? Let us study some familiar examples first.
Convolution Examples: Polynomial multiplication
We have already met such an example in project 1 when we tried to set up the design matrix for a two-dimensional function. This was an example of polynomial multiplication. Let us recast such a problem in terms of the convolution operation. Let us look a the following polynomials to second and third order, respectively:
$$ p(t) = \alpha_0+\alpha_1 t+\alpha_2 t^2, $$and
$$ s(t) = \beta_0+\beta_1 t+\beta_2 t^2+\beta_3 t^3. $$The polynomial multiplication gives us a new polynomial of degree \( 5 \)
$$ z(t) = \delta_0+\delta_1 t+\delta_2 t^2+\delta_3 t^3+\delta_4 t^4+\delta_5 t^5. $$Efficient Polynomial Multiplication
Computing polynomial products can be implemented efficiently if we rewrite the more brute force multiplications using convolution. We note first that the new coefficients are given as
$$ \begin{split} \delta_0=&\alpha_0\beta_0\\ \delta_1=&\alpha_1\beta_0+\alpha_0\beta_1\\ \delta_2=&\alpha_0\beta_2+\alpha_1\beta_1+\alpha_2\beta_0\\ \delta_3=&\alpha_1\beta_2+\alpha_2\beta_1+\alpha_0\beta_3\\ \delta_4=&\alpha_2\beta_2+\alpha_1\beta_3\\ \delta_5=&\alpha_2\beta_3.\\ \end{split} $$We note that \( \alpha_i=0 \) except for \( i\in \left\{0,1,2\right\} \) and \( \beta_i=0 \) except for \( i\in\left\{0,1,2,3\right\} \).
We can then rewrite the coefficients \( \delta_j \) using a discrete convolution as
$$ \delta_j = \sum_{i=-\infty}^{i=\infty}\alpha_i\beta_{j-i}=(\alpha * \beta)_j, $$or as a double sum with restriction \( l=i+j \)
$$ \delta_l = \sum_{ij}\alpha_i\beta_{j}. $$Do you see a potential drawback with these equations?
A more efficient way of coding the above Convolution
Since we only have a finite number of \( \alpha \) and \( \beta \) values which are non-zero, we can rewrite the above convolution expressions as a matrix-vector multiplication
$$ \boldsymbol{\delta}=\begin{bmatrix}\alpha_0 & 0 & 0 & 0 \\ \alpha_1 & \alpha_0 & 0 & 0 \\ \alpha_2 & \alpha_1 & \alpha_0 & 0 \\ 0 & \alpha_2 & \alpha_1 & \alpha_0 \\ 0 & 0 & \alpha_2 & \alpha_1 \\ 0 & 0 & 0 & \alpha_2 \end{bmatrix}\begin{bmatrix} \beta_0 \\ \beta_1 \\ \beta_2 \\ \beta_3\end{bmatrix}. $$The process is commutative and we can easily see that we can rewrite the multiplication in terms of a matrix holding \( \beta \) and a vector holding \( \alpha \). In this case we have
$$ \boldsymbol{\delta}=\begin{bmatrix}\beta_0 & 0 & 0 \\ \beta_1 & \beta_0 & 0 \\ \beta_2 & \beta_1 & \beta_0 \\ \beta_3 & \beta_2 & \beta_1 \\ 0 & \beta_3 & \beta_2 \\ 0 & 0 & \beta_3 \end{bmatrix}\begin{bmatrix} \alpha_0 \\ \alpha_1 \\ \alpha_2\end{bmatrix}. $$Note that the use of these matrices is for mathematical purposes only and not implementation purposes. When implementing the above equation we do not encode (and allocate memory) the matrices explicitely. We rather code the convolutions in the minimal memory footprint that they require.
Does the number of floating point operations change here when we use the commutative property?
Two-dimensional Objects
We often use convolutions over more than one dimension at a time. If we have a two-dimensional image \( I \) as input, we can have a filter defined by a two-dimensional kernel \( K \). This leads to an output \( S \)
$$ S_(i,j)=(I * K)(i,j) = \sum_m\sum_n I(m,n)K(i-m,j-n). $$Convolution is a commutatitave process, which means we can rewrite this equation as
$$ S_(i,j)=(I * K)(i,j) = \sum_m\sum_n I(i-m,j-n)K(m,n). $$Normally the latter is more straightforward to implement in a machine elarning library since there is less variation in the range of values of \( m \) and \( n \).
Cross-Correlation
Many deep learning libraries implement cross-correlation instead of convolution
$$ S_(i,j)=(I * K)(i,j) = \sum_m\sum_n I(i+m,j-+)K(m,n). $$More on Dimensionalities
In feilds like signal processing (and imaging as well), one designs so-called filters. These filters are defined by the convolutions and are often hand-crafted. One may specify filters for smoothing, edge detection, frequency reshaping, and similar operations. However with neural networks the idea is to automatically learn the filters and use many of them in conjunction with non-linear operations (activation functions).
As an example consider a neural network operating on sound sequence data. Assume that we an input vector \( \boldsymbol{x} \) of length \( d=10^6 \). We construct then a neural network with onle hidden layer only with \( 10^4 \) nodes. This means that we will have a weight matrix with \( 10^4\times 10^6=10^{10} \) weights to be determined, together with \( 10^4 \) biases.
Assume furthermore that we have an output layer which is meant to train whether the sound sequence represents a human voice (true) or something else (false). It means that we have only one output node. But since this output node connects to \( 10^4 \) nodes in the hidden layer, there are in total \( 10^4 \) weights to be determined for the output layer, plus one bias. In total we have
$$ \mathrm{NumberParameters}=10^{10}+10^4+10^4+1 \approx 10^{10}, $$that is ten billion parameters to determine.
Further Dimensionality Remarks
In today’s architecture one can train such neural networks, however this is a huge number of parameters for the task at hand. In general, it is a very wasteful and inefficient use of dense matrices as parameters. Just as importantly, such trained network parameters are very specific for the type of input data on which they were trained and the network is not likely to generalize easily to variations in the input.
The main principles that justify convolutions is locality of information and repetion of patterns within the signal. Sound samples of the input in adjacent spots are much more likely to affect each other than those that are very far away. Similarly, sounds are repeated in multiple times in the signal. While slightly simplistic, reasoning about such a sound example demonstrates this. The same principles then apply to images and other similar data.
CNNs in more detail, Lecture from IN5400
CNNs in more detail, building convolutional neural networks in Tensorflow and Keras
As discussed above, CNNs are neural networks built from the assumption that the inputs to the network are 2D images. This is important because the number of features or pixels in images grows very fast with the image size, and an enormous number of weights and biases are needed in order to build an accurate network.
As before, we still have our input, a hidden layer and an output. What's novel about convolutional networks are the convolutional and pooling layers stacked in pairs between the input and the hidden layer. In addition, the data is no longer represented as a 2D feature matrix, instead each input is a number of 2D matrices, typically 1 for each color dimension (Red, Green, Blue).
Setting it up
It means that to represent the entire dataset of images, we require a 4D matrix or tensor. This tensor has the dimensions:
$$ (n_{inputs},\, n_{pixels, width},\, n_{pixels, height},\, depth) . $$The MNIST dataset again
The MNIST dataset consists of grayscale images with a pixel size of \( 28\times 28 \), meaning we require \( 28 \times 28 = 724 \) weights to each neuron in the first hidden layer.
If we were to analyze images of size \( 128\times 128 \) we would require \( 128 \times 128 = 16384 \) weights to each neuron. Even worse if we were dealing with color images, as most images are, we have an image matrix of size \( 128\times 128 \) for each color dimension (Red, Green, Blue), meaning 3 times the number of weights \( = 49152 \) are required for every single neuron in the first hidden layer.
Strong correlations
Images typically have strong local correlations, meaning that a small part of the image varies little from its neighboring regions. If for example we have an image of a blue car, we can roughly assume that a small blue part of the image is surrounded by other blue regions.
Therefore, instead of connecting every single pixel to a neuron in the first hidden layer, as we have previously done with deep neural networks, we can instead connect each neuron to a small part of the image (in all 3 RGB depth dimensions). The size of each small area is fixed, and known as a receptive.
Layers of a CNN
The layers of a convolutional neural network arrange neurons in 3D: width, height and depth. The input image is typically a square matrix of depth 3.
A convolution is performed on the image which outputs a 3D volume of neurons. The weights to the input are arranged in a number of 2D matrices, known as filters.
Each filter slides along the input image, taking the dot product between each small part of the image and the filter, in all depth dimensions. This is then passed through a non-linear function, typically the Rectified Linear (ReLu) function, which serves as the activation of the neurons in the first convolutional layer. This is further passed through a pooling layer, which reduces the size of the convolutional layer, e.g. by taking the maximum or average across some small regions, and this serves as input to the next convolutional layer.
Systematic reduction
By systematically reducing the size of the input volume, through convolution and pooling, the network should create representations of small parts of the input, and then from them assemble representations of larger areas. The final pooling layer is flattened to serve as input to a hidden layer, such that each neuron in the final pooling layer is connected to every single neuron in the hidden layer. This then serves as input to the output layer, e.g. a softmax output for classification.
Prerequisites: Collect and pre-process data
# import necessary packages
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
# ensure the same random numbers appear every time
np.random.seed(0)
# display images in notebook
%matplotlib inline
plt.rcParams['figure.figsize'] = (12,12)
# download MNIST dataset
digits = datasets.load_digits()
# define inputs and labels
inputs = digits.images
labels = digits.target
# RGB images have a depth of 3
# our images are grayscale so they should have a depth of 1
inputs = inputs[:,:,:,np.newaxis]
print("inputs = (n_inputs, pixel_width, pixel_height, depth) = " + str(inputs.shape))
print("labels = (n_inputs) = " + str(labels.shape))
# choose some random images to display
n_inputs = len(inputs)
indices = np.arange(n_inputs)
random_indices = np.random.choice(indices, size=5)
for i, image in enumerate(digits.images[random_indices]):
plt.subplot(1, 5, i+1)
plt.axis('off')
plt.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')
plt.title("Label: %d" % digits.target[random_indices[i]])
plt.show()
Importing Keras and Tensorflow
from tensorflow.keras import datasets, layers, models
from tensorflow.keras.layers import Input
from tensorflow.keras.models import Sequential #This allows appending layers to existing models
from tensorflow.keras.layers import Dense #This allows defining the characteristics of a particular layer
from tensorflow.keras import optimizers #This allows using whichever optimiser we want (sgd,adam,RMSprop)
from tensorflow.keras import regularizers #This allows using whichever regularizer we want (l1,l2,l1_l2)
from tensorflow.keras.utils import to_categorical #This allows using categorical cross entropy as the cost function
#from tensorflow.keras import Conv2D
#from tensorflow.keras import MaxPooling2D
#from tensorflow.keras import Flatten
from sklearn.model_selection import train_test_split
# representation of labels
labels = to_categorical(labels)
# split into train and test data
# one-liner from scikit-learn library
train_size = 0.8
test_size = 1 - train_size
X_train, X_test, Y_train, Y_test = train_test_split(inputs, labels, train_size=train_size,
test_size=test_size)
Running with Keras
def create_convolutional_neural_network_keras(input_shape, receptive_field,
n_filters, n_neurons_connected, n_categories,
eta, lmbd):
model = Sequential()
model.add(layers.Conv2D(n_filters, (receptive_field, receptive_field), input_shape=input_shape, padding='same',
activation='relu', kernel_regularizer=regularizers.l2(lmbd)))
model.add(layers.MaxPooling2D(pool_size=(2, 2)))
model.add(layers.Flatten())
model.add(layers.Dense(n_neurons_connected, activation='relu', kernel_regularizer=regularizers.l2(lmbd)))
model.add(layers.Dense(n_categories, activation='softmax', kernel_regularizer=regularizers.l2(lmbd)))
sgd = optimizers.SGD(lr=eta)
model.compile(loss='categorical_crossentropy', optimizer=sgd, metrics=['accuracy'])
return model
epochs = 100
batch_size = 100
input_shape = X_train.shape[1:4]
receptive_field = 3
n_filters = 10
n_neurons_connected = 50
n_categories = 10
eta_vals = np.logspace(-5, 1, 7)
lmbd_vals = np.logspace(-5, 1, 7)
Final part
CNN_keras = np.zeros((len(eta_vals), len(lmbd_vals)), dtype=object)
for i, eta in enumerate(eta_vals):
for j, lmbd in enumerate(lmbd_vals):
CNN = create_convolutional_neural_network_keras(input_shape, receptive_field,
n_filters, n_neurons_connected, n_categories,
eta, lmbd)
CNN.fit(X_train, Y_train, epochs=epochs, batch_size=batch_size, verbose=0)
scores = CNN.evaluate(X_test, Y_test)
CNN_keras[i][j] = CNN
print("Learning rate = ", eta)
print("Lambda = ", lmbd)
print("Test accuracy: %.3f" % scores[1])
print()
Final visualization
# visual representation of grid search
# uses seaborn heatmap, could probably do this in matplotlib
import seaborn as sns
sns.set()
train_accuracy = np.zeros((len(eta_vals), len(lmbd_vals)))
test_accuracy = np.zeros((len(eta_vals), len(lmbd_vals)))
for i in range(len(eta_vals)):
for j in range(len(lmbd_vals)):
CNN = CNN_keras[i][j]
train_accuracy[i][j] = CNN.evaluate(X_train, Y_train)[1]
test_accuracy[i][j] = CNN.evaluate(X_test, Y_test)[1]
fig, ax = plt.subplots(figsize = (10, 10))
sns.heatmap(train_accuracy, annot=True, ax=ax, cmap="viridis")
ax.set_title("Training Accuracy")
ax.set_ylabel("$\eta$")
ax.set_xlabel("$\lambda$")
plt.show()
fig, ax = plt.subplots(figsize = (10, 10))
sns.heatmap(test_accuracy, annot=True, ax=ax, cmap="viridis")
ax.set_title("Test Accuracy")
ax.set_ylabel("$\eta$")
ax.set_xlabel("$\lambda$")
plt.show()
The CIFAR01 data set
The CIFAR10 dataset contains 60,000 color images in 10 classes, with 6,000 images in each class. The dataset is divided into 50,000 training images and 10,000 testing images. The classes are mutually exclusive and there is no overlap between them.
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
import matplotlib.pyplot as plt
# We import the data set
(train_images, train_labels), (test_images, test_labels) = datasets.cifar10.load_data()
# Normalize pixel values to be between 0 and 1 by dividing by 255.
train_images, test_images = train_images / 255.0, test_images / 255.0
Verifying the data set
To verify that the dataset looks correct, let's plot the first 25 images from the training set and display the class name below each image.
class_names = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
plt.figure(figsize=(10,10))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(train_images[i], cmap=plt.cm.binary)
# The CIFAR labels happen to be arrays,
# which is why you need the extra index
plt.xlabel(class_names[train_labels[i][0]])
plt.show()
Set up the model
The 6 lines of code below define the convolutional base using a common pattern: a stack of Conv2D and MaxPooling2D layers.
As input, a CNN takes tensors of shape (image_height, image_width, color_channels), ignoring the batch size. If you are new to these dimensions, color_channels refers to (R,G,B). In this example, you will configure our CNN to process inputs of shape (32, 32, 3), which is the format of CIFAR images. You can do this by passing the argument input_shape to our first layer.
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(32, 32, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
# Let's display the architecture of our model so far.
model.summary()
You can see that the output of every Conv2D and MaxPooling2D layer is a 3D tensor of shape (height, width, channels). The width and height dimensions tend to shrink as you go deeper in the network. The number of output channels for each Conv2D layer is controlled by the first argument (e.g., 32 or 64). Typically, as the width and height shrink, you can afford (computationally) to add more output channels in each Conv2D layer.
Add Dense layers on top
To complete our model, you will feed the last output tensor from the convolutional base (of shape (4, 4, 64)) into one or more Dense layers to perform classification. Dense layers take vectors as input (which are 1D), while the current output is a 3D tensor. First, you will flatten (or unroll) the 3D output to 1D, then add one or more Dense layers on top. CIFAR has 10 output classes, so you use a final Dense layer with 10 outputs and a softmax activation.
model.add(layers.Flatten())
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(10))
#Here's the complete architecture of our model.
model.summary()
As you can see, our (4, 4, 64) outputs were flattened into vectors of shape (1024) before going through two dense layers.
Compile and train the model
model.compile(optimizer='adam',
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
history = model.fit(train_images, train_labels, epochs=10,
validation_data=(test_images, test_labels))
Finally, evaluate the model
plt.plot(history.history['accuracy'], label='accuracy')
plt.plot(history.history['val_accuracy'], label = 'val_accuracy')
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.ylim([0.5, 1])
plt.legend(loc='lower right')
test_loss, test_acc = model.evaluate(test_images, test_labels, verbose=2)
print(test_acc)
Recurrent neural networks: Overarching view
Till now our focus has been, including convolutional neural networks as well, on feedforward neural networks. The output or the activations flow only in one direction, from the input layer to the output layer.
A recurrent neural network (RNN) looks very much like a feedforward neural network, except that it also has connections pointing backward.
RNNs are used to analyze time series data such as stock prices, and tell you when to buy or sell. In autonomous driving systems, they can anticipate car trajectories and help avoid accidents. More generally, they can work on sequences of arbitrary lengths, rather than on fixed-sized inputs like all the nets we have discussed so far. For example, they can take sentences, documents, or audio samples as input, making them extremely useful for natural language processing systems such as automatic translation and speech-to-text.
Set up of an RNN
See handwritten notes for week 43 and Lectures from CS231 at Stanford
A simple example
# Start importing packages
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
from tensorflow.keras.layers import Input
from tensorflow.keras.models import Model, Sequential
from tensorflow.keras.layers import Dense, SimpleRNN, LSTM, GRU
from tensorflow.keras import optimizers
from tensorflow.keras import regularizers
from tensorflow.keras.utils import to_categorical
# convert into dataset matrix
def convertToMatrix(data, step):
X, Y =[], []
for i in range(len(data)-step):
d=i+step
X.append(data[i:d,])
Y.append(data[d,])
return np.array(X), np.array(Y)
step = 4
N = 1000
Tp = 800
t=np.arange(0,N)
x=np.sin(0.02*t)+2*np.random.rand(N)
df = pd.DataFrame(x)
df.head()
plt.plot(df)
plt.show()
values=df.values
train,test = values[0:Tp,:], values[Tp:N,:]
# add step elements into train and test
test = np.append(test,np.repeat(test[-1,],step))
train = np.append(train,np.repeat(train[-1,],step))
trainX,trainY =convertToMatrix(train,step)
testX,testY =convertToMatrix(test,step)
trainX = np.reshape(trainX, (trainX.shape[0], 1, trainX.shape[1]))
testX = np.reshape(testX, (testX.shape[0], 1, testX.shape[1]))
model = Sequential()
model.add(SimpleRNN(units=32, input_shape=(1,step), activation="relu"))
model.add(Dense(8, activation="relu"))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='rmsprop')
model.summary()
model.fit(trainX,trainY, epochs=100, batch_size=16, verbose=2)
trainPredict = model.predict(trainX)
testPredict= model.predict(testX)
predicted=np.concatenate((trainPredict,testPredict),axis=0)
trainScore = model.evaluate(trainX, trainY, verbose=0)
print(trainScore)
df = pd.DataFrame(x)
pred = pd.DataFrame(predicted)
plt.plot(df,c="b")
plt.plot(pred,c="r")
plt.show()
An extrapolation example
The following code provides an example of how recurrent neural networks can be used to extrapolate to unknown values of physics data sets. Specifically, the data sets used in this program come from a quantum mechanical many-body calculation of energies as functions of the number of particles.
# For matrices and calculations
import numpy as np
# For machine learning (backend for keras)
import tensorflow as tf
# User-friendly machine learning library
# Front end for TensorFlow
import tensorflow.keras
# Different methods from Keras needed to create an RNN
# This is not necessary but it shortened function calls
# that need to be used in the code.
from tensorflow.keras import datasets, layers, models
from tensorflow.keras.layers import Input
from tensorflow.keras import regularizers
from tensorflow.keras.models import Model, Sequential
from tensorflow.keras.layers import Dense, SimpleRNN, LSTM, GRU
# For timing the code
from timeit import default_timer as timer
# For plotting
import matplotlib.pyplot as plt
# The data set
datatype='VaryDimension'
X_tot = np.arange(2, 42, 2)
y_tot = np.array([-0.03077640549, -0.08336233266, -0.1446729567, -0.2116753732, -0.2830637392, -0.3581341341, -0.436462435, -0.5177783846,
-0.6019067271, -0.6887363571, -0.7782028952, -0.8702784034, -0.9649652536, -1.062292565, -1.16231451,
-1.265109911, -1.370782966, -1.479465113, -1.591317992, -1.70653767])
Formatting the Data
The way the recurrent neural networks are trained in this program differs from how machine learning algorithms are usually trained. Typically a machine learning algorithm is trained by learning the relationship between the \( x \) data and the \( y \) data. In this program, the recurrent neural network will be trained to recognize the relationship in a sequence of \( y \) values. This is type of data formatting is typically used for time series forecasting, but it can also be used in any extrapolation (time series forecasting is just a specific type of extrapolation along the time axis). This method of data formatting does not use the \( x \) data and assumes that the \( y \) data are evenly spaced.
For a standard machine learning algorithm, the training data has the form of \( (x,y) \) so the machine learning algorithm learns to associate a \( y \) value with a given \( x \) value. This is useful when the test data has \( x \) values within the same range as the training data. However, for this application, the x values of the test data are outside of the x values of the training data and the traditional method of training a machine learning algorithm does not work as well. For this reason, the recurrent neural network is trained on sequences of y values of the form \( ((y1, y2), y3) \), so that the network is concerned with learning the pattern of the y data and not the relation between the x and y data. As long as the pattern of y data outside of the training region stays relatively stable compared to what was inside the training region, this method of training can produce accurate extrapolations to y values far removed from the training data set.
# FORMAT_DATA
def format_data(data, length_of_sequence = 2):
"""
Inputs:
data(a numpy array): the data that will be the inputs to the recurrent neural
network
length_of_sequence (an int): the number of elements in one iteration of the
sequence patter. For a function approximator use length_of_sequence = 2.
Returns:
rnn_input (a 3D numpy array): the input data for the recurrent neural network. Its
dimensions are length of data - length of sequence, length of sequence,
dimnsion of data
rnn_output (a numpy array): the training data for the neural network
Formats data to be used in a recurrent neural network.
"""
X, Y = [], []
for i in range(len(data)-length_of_sequence):
# Get the next length_of_sequence elements
a = data[i:i+length_of_sequence]
# Get the element that immediately follows that
b = data[i+length_of_sequence]
# Reshape so that each data point is contained in its own array
a = np.reshape (a, (len(a), 1))
X.append(a)
Y.append(b)
rnn_input = np.array(X)
rnn_output = np.array(Y)
return rnn_input, rnn_output
# ## Defining the Recurrent Neural Network Using Keras
#
# The following method defines a simple recurrent neural network in keras consisting of one input layer, one hidden layer, and one output layer.
def rnn(length_of_sequences, batch_size = None, stateful = False):
"""
Inputs:
length_of_sequences (an int): the number of y values in "x data". This is determined
when the data is formatted
batch_size (an int): Default value is None. See Keras documentation of SimpleRNN.
stateful (a boolean): Default value is False. See Keras documentation of SimpleRNN.
Returns:
model (a Keras model): The recurrent neural network that is built and compiled by this
method
Builds and compiles a recurrent neural network with one hidden layer and returns the model.
"""
# Number of neurons in the input and output layers
in_out_neurons = 1
# Number of neurons in the hidden layer
hidden_neurons = 200
# Define the input layer
inp = Input(batch_shape=(batch_size,
length_of_sequences,
in_out_neurons))
# Define the hidden layer as a simple RNN layer with a set number of neurons and add it to
# the network immediately after the input layer
rnn = SimpleRNN(hidden_neurons,
return_sequences=False,
stateful = stateful,
name="RNN")(inp)
# Define the output layer as a dense neural network layer (standard neural network layer)
#and add it to the network immediately after the hidden layer.
dens = Dense(in_out_neurons,name="dense")(rnn)
# Create the machine learning model starting with the input layer and ending with the
# output layer
model = Model(inputs=[inp],outputs=[dens])
# Compile the machine learning model using the mean squared error function as the loss
# function and an Adams optimizer.
model.compile(loss="mean_squared_error", optimizer="adam")
return model
Predicting New Points With A Trained Recurrent Neural Network
def test_rnn (x1, y_test, plot_min, plot_max):
"""
Inputs:
x1 (a list or numpy array): The complete x component of the data set
y_test (a list or numpy array): The complete y component of the data set
plot_min (an int or float): the smallest x value used in the training data
plot_max (an int or float): the largest x valye used in the training data
Returns:
None.
Uses a trained recurrent neural network model to predict future points in the
series. Computes the MSE of the predicted data set from the true data set, saves
the predicted data set to a csv file, and plots the predicted and true data sets w
while also displaying the data range used for training.
"""
# Add the training data as the first dim points in the predicted data array as these
# are known values.
y_pred = y_test[:dim].tolist()
# Generate the first input to the trained recurrent neural network using the last two
# points of the training data. Based on how the network was trained this means that it
# will predict the first point in the data set after the training data. All of the
# brackets are necessary for Tensorflow.
next_input = np.array([[[y_test[dim-2]], [y_test[dim-1]]]])
# Save the very last point in the training data set. This will be used later.
last = [y_test[dim-1]]
# Iterate until the complete data set is created.
for i in range (dim, len(y_test)):
# Predict the next point in the data set using the previous two points.
next = model.predict(next_input)
# Append just the number of the predicted data set
y_pred.append(next[0][0])
# Create the input that will be used to predict the next data point in the data set.
next_input = np.array([[last, next[0]]], dtype=np.float64)
last = next
# Print the mean squared error between the known data set and the predicted data set.
print('MSE: ', np.square(np.subtract(y_test, y_pred)).mean())
# Save the predicted data set as a csv file for later use
name = datatype + 'Predicted'+str(dim)+'.csv'
np.savetxt(name, y_pred, delimiter=',')
# Plot the known data set and the predicted data set. The red box represents the region that was used
# for the training data.
fig, ax = plt.subplots()
ax.plot(x1, y_test, label="true", linewidth=3)
ax.plot(x1, y_pred, 'g-.',label="predicted", linewidth=4)
ax.legend()
# Created a red region to represent the points used in the training data.
ax.axvspan(plot_min, plot_max, alpha=0.25, color='red')
plt.show()
# Check to make sure the data set is complete
assert len(X_tot) == len(y_tot)
# This is the number of points that will be used in as the training data
dim=12
# Separate the training data from the whole data set
X_train = X_tot[:dim]
y_train = y_tot[:dim]
# Generate the training data for the RNN, using a sequence of 2
rnn_input, rnn_training = format_data(y_train, 2)
# Create a recurrent neural network in Keras and produce a summary of the
# machine learning model
model = rnn(length_of_sequences = rnn_input.shape[1])
model.summary()
# Start the timer. Want to time training+testing
start = timer()
# Fit the model using the training data genenerated above using 150 training iterations and a 5%
# validation split. Setting verbose to True prints information about each training iteration.
hist = model.fit(rnn_input, rnn_training, batch_size=None, epochs=150,
verbose=True,validation_split=0.05)
for label in ["loss","val_loss"]:
plt.plot(hist.history[label],label=label)
plt.ylabel("loss")
plt.xlabel("epoch")
plt.title("The final validation loss: {}".format(hist.history["val_loss"][-1]))
plt.legend()
plt.show()
# Use the trained neural network to predict more points of the data set
test_rnn(X_tot, y_tot, X_tot[0], X_tot[dim-1])
# Stop the timer and calculate the total time needed.
end = timer()
print('Time: ', end-start)
Other Things to Try
Changing the size of the recurrent neural network and its parameters can drastically change the results you get from the model. The below code takes the simple recurrent neural network from above and adds a second hidden layer, changes the number of neurons in the hidden layer, and explicitly declares the activation function of the hidden layers to be a sigmoid function. The loss function and optimizer can also be changed but are kept the same as the above network. These parameters can be tuned to provide the optimal result from the network. For some ideas on how to improve the performance of a recurrent neural network.
def rnn_2layers(length_of_sequences, batch_size = None, stateful = False):
"""
Inputs:
length_of_sequences (an int): the number of y values in "x data". This is determined
when the data is formatted
batch_size (an int): Default value is None. See Keras documentation of SimpleRNN.
stateful (a boolean): Default value is False. See Keras documentation of SimpleRNN.
Returns:
model (a Keras model): The recurrent neural network that is built and compiled by this
method
Builds and compiles a recurrent neural network with two hidden layers and returns the model.
"""
# Number of neurons in the input and output layers
in_out_neurons = 1
# Number of neurons in the hidden layer, increased from the first network
hidden_neurons = 500
# Define the input layer
inp = Input(batch_shape=(batch_size,
length_of_sequences,
in_out_neurons))
# Create two hidden layers instead of one hidden layer. Explicitly set the activation
# function to be the sigmoid function (the default value is hyperbolic tangent)
rnn1 = SimpleRNN(hidden_neurons,
return_sequences=True, # This needs to be True if another hidden layer is to follow
stateful = stateful, activation = 'sigmoid',
name="RNN1")(inp)
rnn2 = SimpleRNN(hidden_neurons,
return_sequences=False, activation = 'sigmoid',
stateful = stateful,
name="RNN2")(rnn1)
# Define the output layer as a dense neural network layer (standard neural network layer)
#and add it to the network immediately after the hidden layer.
dens = Dense(in_out_neurons,name="dense")(rnn2)
# Create the machine learning model starting with the input layer and ending with the
# output layer
model = Model(inputs=[inp],outputs=[dens])
# Compile the machine learning model using the mean squared error function as the loss
# function and an Adams optimizer.
model.compile(loss="mean_squared_error", optimizer="adam")
return model
# Check to make sure the data set is complete
assert len(X_tot) == len(y_tot)
# This is the number of points that will be used in as the training data
dim=12
# Separate the training data from the whole data set
X_train = X_tot[:dim]
y_train = y_tot[:dim]
# Generate the training data for the RNN, using a sequence of 2
rnn_input, rnn_training = format_data(y_train, 2)
# Create a recurrent neural network in Keras and produce a summary of the
# machine learning model
model = rnn_2layers(length_of_sequences = 2)
model.summary()
# Start the timer. Want to time training+testing
start = timer()
# Fit the model using the training data genenerated above using 150 training iterations and a 5%
# validation split. Setting verbose to True prints information about each training iteration.
hist = model.fit(rnn_input, rnn_training, batch_size=None, epochs=150,
verbose=True,validation_split=0.05)
# This section plots the training loss and the validation loss as a function of training iteration.
# This is not required for analyzing the couple cluster data but can help determine if the network is
# being overtrained.
for label in ["loss","val_loss"]:
plt.plot(hist.history[label],label=label)
plt.ylabel("loss")
plt.xlabel("epoch")
plt.title("The final validation loss: {}".format(hist.history["val_loss"][-1]))
plt.legend()
plt.show()
# Use the trained neural network to predict more points of the data set
test_rnn(X_tot, y_tot, X_tot[0], X_tot[dim-1])
# Stop the timer and calculate the total time needed.
end = timer()
print('Time: ', end-start)
Other Types of Recurrent Neural Networks
Besides a simple recurrent neural network layer, there are two other commonly used types of recurrent neural network layers: Long Short Term Memory (LSTM) and Gated Recurrent Unit (GRU). For a short introduction to these layers see https://medium.com/mindboard/lstm-vs-gru-experimental-comparison-955820c21e8b and https://medium.com/mindboard/lstm-vs-gru-experimental-comparison-955820c21e8b.
The first network created below is similar to the previous network, but it replaces the SimpleRNN layers with LSTM layers. The second network below has two hidden layers made up of GRUs, which are preceeded by two dense (feeddorward) neural network layers. These dense layers "preprocess" the data before it reaches the recurrent layers. This architecture has been shown to improve the performance of recurrent neural networks (see the link above and also https://arxiv.org/pdf/1807.02857.pdf.
def lstm_2layers(length_of_sequences, batch_size = None, stateful = False):
"""
Inputs:
length_of_sequences (an int): the number of y values in "x data". This is determined
when the data is formatted
batch_size (an int): Default value is None. See Keras documentation of SimpleRNN.
stateful (a boolean): Default value is False. See Keras documentation of SimpleRNN.
Returns:
model (a Keras model): The recurrent neural network that is built and compiled by this
method
Builds and compiles a recurrent neural network with two LSTM hidden layers and returns the model.
"""
# Number of neurons on the input/output layer and the number of neurons in the hidden layer
in_out_neurons = 1
hidden_neurons = 250
# Input Layer
inp = Input(batch_shape=(batch_size,
length_of_sequences,
in_out_neurons))
# Hidden layers (in this case they are LSTM layers instead if SimpleRNN layers)
rnn= LSTM(hidden_neurons,
return_sequences=True,
stateful = stateful,
name="RNN", use_bias=True, activation='tanh')(inp)
rnn1 = LSTM(hidden_neurons,
return_sequences=False,
stateful = stateful,
name="RNN1", use_bias=True, activation='tanh')(rnn)
# Output layer
dens = Dense(in_out_neurons,name="dense")(rnn1)
# Define the midel
model = Model(inputs=[inp],outputs=[dens])
# Compile the model
model.compile(loss='mean_squared_error', optimizer='adam')
# Return the model
return model
def dnn2_gru2(length_of_sequences, batch_size = None, stateful = False):
"""
Inputs:
length_of_sequences (an int): the number of y values in "x data". This is determined
when the data is formatted
batch_size (an int): Default value is None. See Keras documentation of SimpleRNN.
stateful (a boolean): Default value is False. See Keras documentation of SimpleRNN.
Returns:
model (a Keras model): The recurrent neural network that is built and compiled by this
method
Builds and compiles a recurrent neural network with four hidden layers (two dense followed by
two GRU layers) and returns the model.
"""
# Number of neurons on the input/output layers and hidden layers
in_out_neurons = 1
hidden_neurons = 250
# Input layer
inp = Input(batch_shape=(batch_size,
length_of_sequences,
in_out_neurons))
# Hidden Dense (feedforward) layers
dnn = Dense(hidden_neurons/2, activation='relu', name='dnn')(inp)
dnn1 = Dense(hidden_neurons/2, activation='relu', name='dnn1')(dnn)
# Hidden GRU layers
rnn1 = GRU(hidden_neurons,
return_sequences=True,
stateful = stateful,
name="RNN1", use_bias=True)(dnn1)
rnn = GRU(hidden_neurons,
return_sequences=False,
stateful = stateful,
name="RNN", use_bias=True)(rnn1)
# Output layer
dens = Dense(in_out_neurons,name="dense")(rnn)
# Define the model
model = Model(inputs=[inp],outputs=[dens])
# Compile the mdoel
model.compile(loss='mean_squared_error', optimizer='adam')
# Return the model
return model
# Check to make sure the data set is complete
assert len(X_tot) == len(y_tot)
# This is the number of points that will be used in as the training data
dim=12
# Separate the training data from the whole data set
X_train = X_tot[:dim]
y_train = y_tot[:dim]
# Generate the training data for the RNN, using a sequence of 2
rnn_input, rnn_training = format_data(y_train, 2)
# Create a recurrent neural network in Keras and produce a summary of the
# machine learning model
# Change the method name to reflect which network you want to use
model = dnn2_gru2(length_of_sequences = 2)
model.summary()
# Start the timer. Want to time training+testing
start = timer()
# Fit the model using the training data genenerated above using 150 training iterations and a 5%
# validation split. Setting verbose to True prints information about each training iteration.
hist = model.fit(rnn_input, rnn_training, batch_size=None, epochs=150,
verbose=True,validation_split=0.05)
# This section plots the training loss and the validation loss as a function of training iteration.
# This is not required for analyzing the couple cluster data but can help determine if the network is
# being overtrained.
for label in ["loss","val_loss"]:
plt.plot(hist.history[label],label=label)
plt.ylabel("loss")
plt.xlabel("epoch")
plt.title("The final validation loss: {}".format(hist.history["val_loss"][-1]))
plt.legend()
plt.show()
# Use the trained neural network to predict more points of the data set
test_rnn(X_tot, y_tot, X_tot[0], X_tot[dim-1])
# Stop the timer and calculate the total time needed.
end = timer()
print('Time: ', end-start)
# ### Training Recurrent Neural Networks in the Standard Way (i.e. learning the relationship between the X and Y data)
#
# Finally, comparing the performace of a recurrent neural network using the standard data formatting to the performance of the network with time sequence data formatting shows the benefit of this type of data formatting with extrapolation.
# Check to make sure the data set is complete
assert len(X_tot) == len(y_tot)
# This is the number of points that will be used in as the training data
dim=12
# Separate the training data from the whole data set
X_train = X_tot[:dim]
y_train = y_tot[:dim]
# Reshape the data for Keras specifications
X_train = X_train.reshape((dim, 1))
y_train = y_train.reshape((dim, 1))
# Create a recurrent neural network in Keras and produce a summary of the
# machine learning model
# Set the sequence length to 1 for regular data formatting
model = rnn(length_of_sequences = 1)
model.summary()
# Start the timer. Want to time training+testing
start = timer()
# Fit the model using the training data genenerated above using 150 training iterations and a 5%
# validation split. Setting verbose to True prints information about each training iteration.
hist = model.fit(X_train, y_train, batch_size=None, epochs=150,
verbose=True,validation_split=0.05)
# This section plots the training loss and the validation loss as a function of training iteration.
# This is not required for analyzing the couple cluster data but can help determine if the network is
# being overtrained.
for label in ["loss","val_loss"]:
plt.plot(hist.history[label],label=label)
plt.ylabel("loss")
plt.xlabel("epoch")
plt.title("The final validation loss: {}".format(hist.history["val_loss"][-1]))
plt.legend()
plt.show()
# Use the trained neural network to predict the remaining data points
X_pred = X_tot[dim:]
X_pred = X_pred.reshape((len(X_pred), 1))
y_model = model.predict(X_pred)
y_pred = np.concatenate((y_tot[:dim], y_model.flatten()))
# Plot the known data set and the predicted data set. The red box represents the region that was used
# for the training data.
fig, ax = plt.subplots()
ax.plot(X_tot, y_tot, label="true", linewidth=3)
ax.plot(X_tot, y_pred, 'g-.',label="predicted", linewidth=4)
ax.legend()
# Created a red region to represent the points used in the training data.
ax.axvspan(X_tot[0], X_tot[dim], alpha=0.25, color='red')
plt.show()
# Stop the timer and calculate the total time needed.
end = timer()
print('Time: ', end-start)
Generative Models
Generative models describe a class of statistical models that are a contrast to discriminative models. Informally we say that generative models can generate new data instances while discriminative models discriminate between different kinds of data instances. A generative model could generate new photos of animals that look like 'real' animals while a discriminative model could tell a dog from a cat. More formally, given a data set \( x \) and a set of labels / targets \( y \). Generative models capture the joint probability \( p(x, y) \), or just \( p(x) \) if there are no labels, while discriminative models capture the conditional probability \( p(y | x) \). Discriminative models generally try to draw boundaries in the data space (often high dimensional), while generative models try to model how data is placed throughout the space.
Note: this material is thanks to Linus Ekstrøm.
Generative Adversarial Networks
Generative Adversarial Networks are a type of unsupervised machine learning algorithm proposed by Goodfellow et. al in 2014 (short and good article).
The simplest formulation of the model is based on a game theoretic approach, zero sum game, where we pit two neural networks against one another. We define two rival networks, one generator \( g \), and one discriminator \( d \). The generator directly produces samples
$$ \begin{equation} x = g(z; \theta^{(g)}) \label{_auto1} \end{equation} $$Discriminator
The discriminator attempts to distinguish between samples drawn from the training data and samples drawn from the generator. In other words, it tries to tell the difference between the fake data produced by \( g \) and the actual data samples we want to do prediction on. The discriminator outputs a probability value given by
$$ \begin{equation} d(x; \theta^{(d)}) \label{_auto2} \end{equation} $$indicating the probability that \( x \) is a real training example rather than a fake sample the generator has generated. The simplest way to formulate the learning process in a generative adversarial network is a zero-sum game, in which a function
$$ \begin{equation} v(\theta^{(g)}, \theta^{(d)}) \label{_auto3} \end{equation} $$determines the reward for the discriminator, while the generator gets the conjugate reward
$$ \begin{equation} -v(\theta^{(g)}, \theta^{(d)}) \label{_auto4} \end{equation} $$Learning Process
During learning both of the networks maximize their own reward function, so that the generator gets better and better at tricking the discriminator, while the discriminator gets better and better at telling the difference between the fake and real data. The generator and discriminator alternate on which one trains at one time (i.e. for one epoch). In other words, we keep the generator constant and train the discriminator, then we keep the discriminator constant to train the generator and repeat. It is this back and forth dynamic which lets GANs tackle otherwise intractable generative problems. As the generator improves with training, the discriminator's performance gets worse because it cannot easily tell the difference between real and fake. If the generator ends up succeeding perfectly, the the discriminator will do no better than random guessing i.e. 50\%. This progression in the training poses a problem for the convergence criteria for GANs. The discriminator feedback gets less meaningful over time, if we continue training after this point then the generator is effectively training on junk data which can undo the learning up to that point. Therefore, we stop training when the discriminator starts outputting \( 1/2 \) everywhere.
More about the Learning Process
At convergence we have
$$ \begin{equation} g^* = \underset{g}{\mathrm{argmin}}\hspace{2pt} \underset{d}{\mathrm{max}}v(\theta^{(g)}, \theta^{(d)}) \label{_auto5} \end{equation} $$The default choice for \( v \) is
$$ \begin{equation} v(\theta^{(g)}, \theta^{(d)}) = \mathbb{E}_{x\sim p_\mathrm{data}}\log d(x) + \mathbb{E}_{x\sim p_\mathrm{model}} \log (1 - d(x)) \label{_auto6} \end{equation} $$The main motivation for the design of GANs is that the learning process requires neither approximate inference (variational autoencoders for example) nor approximation of a partition function. In the case where
$$ \begin{equation} \underset{d}{\mathrm{max}}v(\theta^{(g)}, \theta^{(d)}) \label{_auto7} \end{equation} $$is convex in $\theta^{(g)} then the procedure is guaranteed to converge and is asymptotically consistent ( Seth Lloyd on QuGANs ).
Additional References
This is in general not the case and it is possible to get situations where the training process never converges because the generator and discriminator chase one another around in the parameter space indefinitely. A much deeper discussion on the currently open research problem of GAN convergence is available here. To anyone interested in learning more about GANs it is a highly recommended read. Direct quote: "In this best-performing formulation, the generator aims to increase the log probability that the discriminator makes a mistake, rather than aiming to decrease the log probability that the discriminator makes the correct prediction." Another interesting read
Writing Our First Generative Adversarial Network
Let us now move on to actually implementing a GAN in tensorflow. We will study the performance of our GAN on the MNIST dataset. This code is based on and adapted from the google tutorial
First we import our libraries
import os
import time
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from tensorflow.keras import layers
from tensorflow.keras.utils import plot_model
Next we define our hyperparameters and import our data the usual way
BUFFER_SIZE = 60000
BATCH_SIZE = 256
EPOCHS = 30
data = tf.keras.datasets.mnist.load_data()
(train_images, train_labels), (test_images, test_labels) = data
train_images = np.reshape(train_images, (train_images.shape[0],
28,
28,
1)).astype('float32')
# we normalize between -1 and 1
train_images = (train_images - 127.5) / 127.5
training_dataset = tf.data.Dataset.from_tensor_slices(
train_images).shuffle(BUFFER_SIZE).batch(BATCH_SIZE)
MNIST and GANs
Let's have a quick look
plt.imshow(train_images[0], cmap='Greys')
plt.show()
Now we define our two models. This is where the 'magic' happens. There are a huge amount of possible formulations for both models. A lot of engineering and trial and error can be done here to try to produce better performing models. For more advanced GANs this is by far the step where you can 'make or break' a model.
We start with the generator. As stated in the introductory text the generator \( g \) upsamples from a random sample to the shape of what we want to predict. In our case we are trying to predict MNIST images (\( 28\times 28 \) pixels).
def generator_model():
"""
The generator uses upsampling layers tf.keras.layers.Conv2DTranspose() to
produce an image from a random seed. We start with a Dense layer taking this
random sample as an input and subsequently upsample through multiple
convolutional layers.
"""
# we define our model
model = tf.keras.Sequential()
# adding our input layer. Dense means that every neuron is connected and
# the input shape is the shape of our random noise. The units need to match
# in some sense the upsampling strides to reach our desired output shape.
# we are using 100 random numbers as our seed
model.add(layers.Dense(units=7*7*BATCH_SIZE,
use_bias=False,
input_shape=(100, )))
# we normalize the output form the Dense layer
model.add(layers.BatchNormalization())
# and add an activation function to our 'layer'. LeakyReLU avoids vanishing
# gradient problem
model.add(layers.LeakyReLU())
model.add(layers.Reshape((7, 7, BATCH_SIZE)))
assert model.output_shape == (None, 7, 7, BATCH_SIZE)
# even though we just added four keras layers we think of everything above
# as 'one' layer
# next we add our upscaling convolutional layers
model.add(layers.Conv2DTranspose(filters=128,
kernel_size=(5, 5),
strides=(1, 1),
padding='same',
use_bias=False))
model.add(layers.BatchNormalization())
model.add(layers.LeakyReLU())
assert model.output_shape == (None, 7, 7, 128)
model.add(layers.Conv2DTranspose(filters=64,
kernel_size=(5, 5),
strides=(2, 2),
padding='same',
use_bias=False))
model.add(layers.BatchNormalization())
model.add(layers.LeakyReLU())
assert model.output_shape == (None, 14, 14, 64)
model.add(layers.Conv2DTranspose(filters=1,
kernel_size=(5, 5),
strides=(2, 2),
padding='same',
use_bias=False,
activation='tanh'))
assert model.output_shape == (None, 28, 28, 1)
return model
And there we have our 'simple' generator model. Now we move on to defining our discriminator model \( d \), which is a convolutional neural network based image classifier.
def discriminator_model():
"""
The discriminator is a convolutional neural network based image classifier
"""
# we define our model
model = tf.keras.Sequential()
model.add(layers.Conv2D(filters=64,
kernel_size=(5, 5),
strides=(2, 2),
padding='same',
input_shape=[28, 28, 1]))
model.add(layers.LeakyReLU())
# adding a dropout layer as you do in conv-nets
model.add(layers.Dropout(0.3))
model.add(layers.Conv2D(filters=128,
kernel_size=(5, 5),
strides=(2, 2),
padding='same'))
model.add(layers.LeakyReLU())
# adding a dropout layer as you do in conv-nets
model.add(layers.Dropout(0.3))
model.add(layers.Flatten())
model.add(layers.Dense(1))
return model
Other Models
Let us take a look at our models. Note: double click images for bigger view.
generator = generator_model()
plot_model(generator, show_shapes=True, rankdir='LR')
discriminator = discriminator_model()
plot_model(discriminator, show_shapes=True, rankdir='LR')
Next we need a few helper objects we will use in training
cross_entropy = tf.keras.losses.BinaryCrossentropy(from_logits=True)
generator_optimizer = tf.keras.optimizers.Adam(1e-4)
discriminator_optimizer = tf.keras.optimizers.Adam(1e-4)
The first object, cross_entropy is our loss function and the two others are our optimizers. Notice we use the same learning rate for both \( g \) and \( d \). This is because they need to improve their accuracy at approximately equal speeds to get convergence (not necessarily exactly equal). Now we define our loss functions
def generator_loss(fake_output):
loss = cross_entropy(tf.ones_like(fake_output), fake_output)
return loss
def discriminator_loss(real_output, fake_output):
real_loss = cross_entropy(tf.ones_like(real_output), real_output)
fake_loss = cross_entropy(tf.zeros_liks(fake_output), fake_output)
total_loss = real_loss + fake_loss
return total_loss
Next we define a kind of seed to help us compare the learning process over multiple training epochs.
noise_dimension = 100
n_examples_to_generate = 16
seed_images = tf.random.normal([n_examples_to_generate, noise_dimension])
Training Step
Now we have everything we need to define our training step, which we will apply for every step in our training loop. Notice the @tf.function flag signifying that the function is tensorflow 'compiled'. Removing this flag doubles the computation time.
@tf.function
def train_step(images):
noise = tf.random.normal([BATCH_SIZE, noise_dimension])
with tf.GradientTape() as gen_tape, tf.GradientTape() as disc_tape:
generated_images = generator(noise, training=True)
real_output = discriminator(images, training=True)
fake_output = discriminator(generated_images, training=True)
gen_loss = generator_loss(fake_output)
disc_loss = discriminator_loss(real_output, fake_output)
gradients_of_generator = gen_tape.gradient(gen_loss,
generator.trainable_variables)
gradients_of_discriminator = disc_tape.gradient(disc_loss,
discriminator.trainable_variables)
generator_optimizer.apply_gradients(zip(gradients_of_generator,
generator.trainable_variables))
discriminator_optimizer.apply_gradients(zip(gradients_of_discriminator,
discriminator.trainable_variables))
return gen_loss, disc_loss
Next we define a helper function to produce an output over our training epochs to see the predictive progression of our generator model. Note: I am including this code here, but comment it out in the training loop.
def generate_and_save_images(model, epoch, test_input):
# we're making inferences here
predictions = model(test_input, training=False)
fig = plt.figure(figsize=(4, 4))
for i in range(predictions.shape[0]):
plt.subplot(4, 4, i+1)
plt.imshow(predictions[i, :, :, 0] * 127.5 + 127.5, cmap='gray')
plt.axis('off')
plt.savefig(f'./images_from_seed_images/image_at_epoch_{str(epoch).zfill(3)}.png')
plt.close()
#plt.show()
Checkpoints
Setting up checkpoints to periodically save our model during training so that everything is not lost even if the program were to somehow terminate while training.
# Setting up checkpoints to save model during training
checkpoint_dir = './training_checkpoints'
checkpoint_prefix = os.path.join(checkpoint_dir, 'ckpt')
checkpoint = tf.train.Checkpoint(generator_optimizer=generator_optimizer,
discriminator_optimizer=discriminator_optimizer,
generator=generator,
discriminator=discriminator)
Now we define our training loop
def train(dataset, epochs):
generator_loss_list = []
discriminator_loss_list = []
for epoch in range(epochs):
start = time.time()
for image_batch in dataset:
gen_loss, disc_loss = train_step(image_batch)
generator_loss_list.append(gen_loss.numpy())
discriminator_loss_list.append(disc_loss.numpy())
#generate_and_save_images(generator, epoch + 1, seed_images)
if (epoch + 1) % 15 == 0:
checkpoint.save(file_prefix=checkpoint_prefix)
print(f'Time for epoch {epoch} is {time.time() - start}')
#generate_and_save_images(generator, epochs, seed_images)
loss_file = './data/lossfile.txt'
with open(loss_file, 'w') as outfile:
outfile.write(str(generator_loss_list))
outfile.write('\n')
outfile.write('\n')
outfile.write(str(discriminator_loss_list))
outfile.write('\n')
outfile.write('\n')
To train simply call this function. Warning: this might take a long time so there is a folder of a pretrained network already included in the repository.
train(train_dataset, EPOCHS)
And here is the result of training our model for 100 epochs
Now to avoid having to train and everything, which will take a while depending on your computer setup we now load in the model which produced the above gif.
checkpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))
restored_generator = checkpoint.generator
restored_discriminator = checkpoint.discriminator
print(restored_generator)
print(restored_discriminator)
Exploring the Latent Space
We have successfully loaded in our latest model. Let us now play around a bit and see what kind of things we can learn about this model. Our generator takes an array of 100 numbers. One idea can be to try to systematically change our input. Let us try and see what we get
def generate_latent_points(number=100, scale_means=1, scale_stds=1):
latent_dim = 100
means = scale_means * tf.linspace(-1, 1, num=latent_dim)
stds = scale_stds * tf.linspace(-1, 1, num=latent_dim)
latent_space_value_range = tf.random.normal([number, latent_dim],
means,
stds,
dtype=tf.float64)
return latent_space_value_range
def generate_images(latent_points):
# notice we set training to false because we are making inferences
generated_images = restored_generator.predict(latent_points)
return generated_images
def plot_result(generated_images, number=100):
# obviously this assumes sqrt number is an int
fig, axs = plt.subplots(int(np.sqrt(number)), int(np.sqrt(number)),
figsize=(10, 10))
for i in range(int(np.sqrt(number))):
for j in range(int(np.sqrt(number))):
axs[i, j].imshow(generated_images[i*j], cmap='Greys')
axs[i, j].axis('off')
plt.show()
generated_images = generate_images(generate_latent_points())
plot_result(generated_images)
Getting Results
We see that the generator generates images that look like MNIST numbers: \( 1, 4, 7, 9 \). Let's try to tweak it a bit more to see if we are able to generate a similar plot where we generate every MNIST number. Let us now try to 'move' a bit around in the latent space. Note: decrease the plot number if these following cells take too long to run on your computer.
plot_number = 225
generated_images = generate_images(generate_latent_points(number=plot_number,
scale_means=5,
scale_stds=1))
plot_result(generated_images, number=plot_number)
generated_images = generate_images(generate_latent_points(number=plot_number,
scale_means=-5,
scale_stds=1))
plot_result(generated_images, number=plot_number)
generated_images = generate_images(generate_latent_points(number=plot_number,
scale_means=1,
scale_stds=5))
plot_result(generated_images, number=plot_number)
Again, we have found something interesting. Moving around using our means takes us from digit to digit, while moving around using our standard deviations seem to increase the number of different digits! In the last image above, we can barely make out every MNIST digit. Let us make on last plot using this information by upping the standard deviation of our Gaussian noises.
plot_number = 400
generated_images = generate_images(generate_latent_points(number=plot_number,
scale_means=1,
scale_stds=10))
plot_result(generated_images, number=plot_number)
A pretty cool result! We see that our generator indeed has learned a distribution which qualitatively looks a whole lot like the MNIST dataset.
Interpolating Between MNIST Digits
Another interesting way to explore the latent space of our generator model is by interpolating between the MNIST digits. This section is largely based on this excellent blogpost by Jason Brownlee.
So let us start by defining a function to interpolate between two points in the latent space.
def interpolation(point_1, point_2, n_steps=10):
ratios = np.linspace(0, 1, num=n_steps)
vectors = []
for i, ratio in enumerate(ratios):
vectors.append(((1.0 - ratio) * point_1 + ratio * point_2))
return tf.stack(vectors)
Now we have all we need to do our interpolation analysis.
plot_number = 100
latent_points = generate_latent_points(number=plot_number)
results = None
for i in range(0, 2*np.sqrt(plot_number), 2):
interpolated = interpolation(latent_points[i], latent_points[i+1])
generated_images = generate_images(interpolated)
if results is None:
results = generated_images
else:
results = tf.stack((results, generated_images))
plot_results(results, plot_number)